A Leap Forward in Bioscience
Eric Horvitz, Chief Scientific Officer
Microsoft
Published on November 22, 2021
|
B |
reakthroughs
in the sciences have been powered by tools that advance our ability to see and understand.
In biology, great inflections followed the advent of the optical microscope of Antonie van Leeuwenhoek, the electron microscope, x-ray
crystallography, gene sequencing, and high-dimensional microarrays. I’m excited
about directions forward with the rise of the computational microscope. A leap forward in bioscience,
reported in Science this month, gives
us a glimpse into the possibilities.
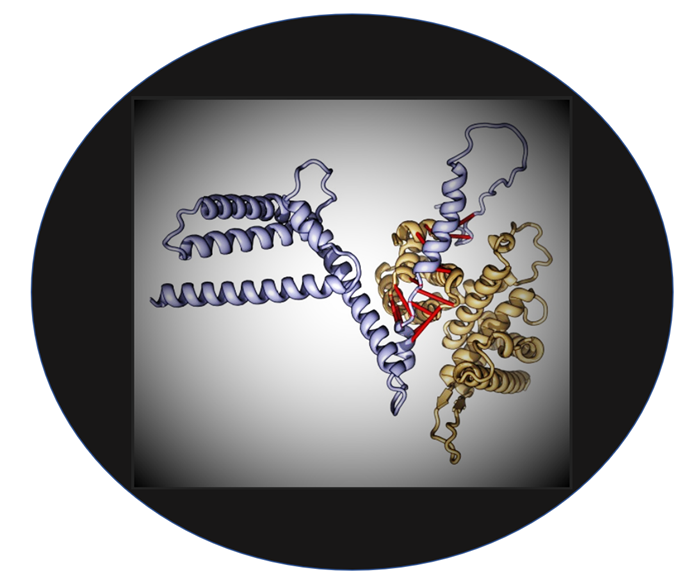
The late-breaking
article by Humphreys et al. marks a
breakthrough in our abilities to peer into the foundations of cellular
functioning. Protein interactions and the complexes formed when proteins come
together are the engines of biological systems. Despite intensive studies on
protein interactions, much remains
unknown about protein interactions and
complexes.
Developing
more comprehensive understandings about protein interactions would help to
decode multiple mysteries of biology and accelerate the creation of novel
therapeutics and cures. The new work shows how AI pipelines can illuminate the dark matter of the
thousands of interactions among proteins and the complexes they form, shining
light on the protein interactome of cells.
The
project on
identifying protein complexes is based at the Institute for Protein Design
(IPD) at the University of Washington (UW), and has been nurtured by an
exciting collaboration
between UW and Microsoft. Microsoft has provided IPD with
large-scale computing resources on Azure and assistance with deep-learning
engineering, and research teams across the groups have been sharing insights,
directions, and data. We have been learning together about the challenging
workloads and aspirations for computing in the biosciences. Our support to date
has included efforts to evolve Rosetta, UW’s protein modeling software suite,
into RoseTTAFold, and the more recent effort on identifying
protein complexes, representing a shift of focus of attention from identifying
protein structure to understanding protein function.
Breakthrough in
identifying protein complexes
In the
results reported in Science, hundreds of previously unknown protein
complexes were identified in one of the simplest eukaryotes, the unicellular Saccharomyces cerevisiae (aka
yeast). The newly discovered complexes have
been linked to a breadth of fundamental processes in eukaryotic cells,
including repairing damage to DNA, doing metabolism, translating RNA into
proteins inside ribosomes, and transporting molecules through cell membranes.
Some of the complexes are employed for tugging chromosomes apart during cell
reproduction, playing a key role in the prominent dynamics of the alignment and
motion of chromosomes during meiosis and mitosis.
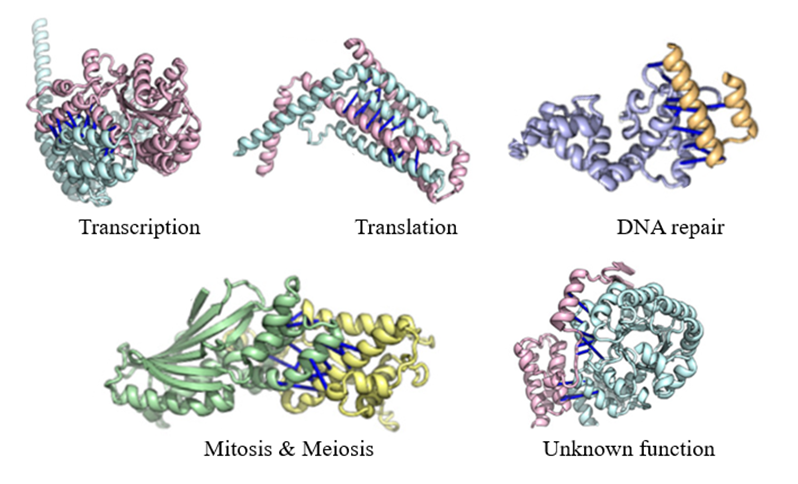
Decoding Protein Complexes. Deep learning methods
have been used to identify likely protein complexes in eukaryotic cells. The
complexes have been linked to processes of transcription, translation, DNA
repair, mitosis and meiosis, metabolism, and protein transport within cells and
across membranes. The dark blue lines indicate likely points of contact
predicted between the proteins. The function of some of the identified
complexes are mysteries (complexes drawn from Humphreys et al. (2021)).
I selected
several examples of the inferred protein complexes to share in the figure
above. The inferences include the structure and configuration of interacting
proteins, as well as predictions about the contact points (dark blue lines)
between proteins participating in the complexes. Beyond pairs, the team
identified complexes of three and four interacting proteins. It is not hard to
imagine how the views of the protein complexes enabled by AI-powered optics can
provide insights about potential drug targets and supercharge drug discovery
efforts.
One of
the protein complexes selected has an intriguing label: unknown function.
The AI methods have helped us to discover protein complexes with roles that we
do not yet understand! Using AI methods to identify mysterious actors on the
cellular stage foreshadows exciting discoveries ahead enabled by our new
computational microscopes.
Leveraging evolutionary signals
The leap
forward with inferring protein complexes builds on the exciting breakthrough
reported last year on using deep neural networks to predict the 3D structure of
proteins from their
underlying amino acid sequences. DeepMind’s AlphaFold system demonstrated how a deep learning pipeline could
predict protein structure as well as x-ray crystallography. The methods harnessed to identify
the previously unknown protein complexes and their configurations leveraged the
capabilities of both AlphaFold and RoseTTAFold.
The analysis of protein complexes in yeast started with a consideration of evolutionary signals about the co-evolution of proteins. The intuition is that proteins that interact in critical ways in a cell would likely have had to evolve together across animal species to maintain their key functions. The team compared the amino acid sequences of 6000 known yeast proteins to their orthologs, the different variants of the proteins appearing in thousands of plants and animals. That analysis led to the identification of 8.3 million pairs of proteins that appear to have changed in synchrony over the millennia.
In a
second step, the likelihood that each of the candidate pairs that form protein
complexes were explored via a modified version of RoseTTAFold
and AlphaFold, yielding 3D structures and potential points of interactions
between proteins. In the end, the computing pipeline identified 1506 proteins
that are likely to interact and the 3D structures of 712 were computed.
New understandings of our cells
The new results are deeply relevant to human biology because the
machinery of our cells is remarkably similar to the
machinery of yeast. We, and all the animals and plants around us, are
constructed from eukaryotic cell building blocks. The eukaryotic cell has
served as a resilient “platform” for the evolution of complex life. The core
structure and processes of eukaryotic cells have largely been conserved; they
have changed surprisingly little over millions of years of evolution across
thousands of plants and animals. So, understanding
the interactions, dalliances, and couplings of proteins within the unicellular
yeast is a leap forward on understanding the functioning of our own cells--and
an important steppingstone on the aspirational path of decoding the full human
protein interactome.
There’s much work ahead on the path to developing a more complete
understanding of the human protein interactome. I’m optimistic that we’re well
on our way and I’m excited about the next steps in our collaborations and learnings.
___________________________________
horvitz@microsoft.com